Viruses have evolved a highly complex and impressive array of interactions with host-cells. One of the ways that viruses “hijack” the host cell and make use of the wider set of cellular machinery available is by enlisting cellular proteins and signaling pathways to create sub-domains in the cell. These sub-domains act as micro-environments within the cells to maximise the production of viral progeny from the infected host cell.
In a recent study “Murine polyomavirus DNA transitions through spatially distinct nuclear replication subdomains during infection” published in PLOS Pathogens, Douglas Peters and Robert Garcea from the Department of Molecular, Cellular, and Developmental Biology and BioFrontiers Institute at the University of Colorado Boulder investigated the sub-nuclear domains termed virus replication centres (VRCs) formed by Polyomavirus (MuPyV). The Polyomaviridae are a group of small DNA viruses that have a wide host range, with some causing infection of humans. Replication of these viruses as well as other DNA viruses, including adenoviruses, herpesviruses and papillomaviruses form within these replication compartments during infection.
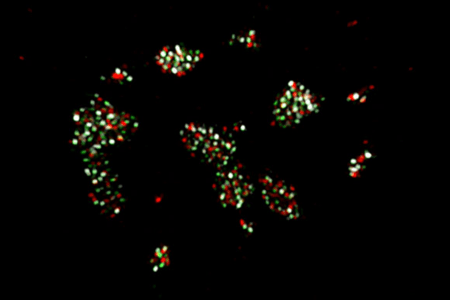
MuPyV infected cells treated with HU (Hydroxyurea) used to investigate organization within virus replication centres (VRC). C57BL/6 mouse embryonic fibroblasts (MEFs) cells were treated with HU then allowed to recover for 4hrs prior to fixation. Ch. 1 (green) is the cellular replication protein A (RPA32) Ch. 2 (white) is vDNA labeled by FISH. Ch. 3 (red) is the viral LT protein. Image courtesy of Douglas Peters, Garcea Lab.
Enhancing resolution within sub-domains using 3D-SIM
Epifluorescence and confocal microscopy are widely used to study many aspects of virus-host cell interactions. The presence of MuPyV virus replication centres has been reported using these techniques. However, it is not possible to resolve any detail of VRCs using such diffraction-limited microscopy. For this, an alternative imaging technique is required. In this study, Peters and Garcea used 3D-SIM to investigate the organization within the VRCs. SIM (Structured Illumination Microscopy) provides up to a two-fold increase in resolution compared to normal fluorescence microscopy. This is achieved by switching between structured illumination patterns, with the interference creating a Moire pattern. Fourier analysis is then applied to the resultant data and images with 100-200 nm resolution can be attained. 3D-SIM, as used in this study, is a development of SIM that uses illumination sequences that are arranged in a series of 3D spatial pattern to allow for higher spatial resolution in 3D.
Understanding the Organizational Dynamics within VRCs
The MuPyV genome codes for six proteins. Three of these viral proteins are early “tumour” expression, T-antigens. The large (LT), middle (MT), and small (ST) T-antigens. These have been linked to a range of functions and interact with cellular proteins during infection. The LT protein is found localized within the nucleus at the VRCs, binding to the viral origin of replication and has a helicase function. A number of host cell DNA replication factors, such as replication protein A (RPA) and DNA polymerase δ, have also been found to be localized to VRC, as have DNA damage response proteins (DDRs).
In this study, Peters and Garcea reported that they were able to track components of the VRCs such as RPA, the viral LT and viral DNA and observe their organization in at least 2 separate spatially and functionally distinct subdomains. A “replication-associated” subdomain as defined by bright LT and dim RPA32 signals. The second subdomain identified was a “repair-associated” subdomain with focal RPA32, pRPA32, pATM localization and the accumulation of vDNA. They were also able to observe that post-synthesis, the viral DNA was being sequentially associated with these domains within the nucleus.
These results show the benefits of the 3D-SIM technique when compared to diffraction-limited microscopy techniques. Further work should help provide insight into the trafficking of the viral genomes during replication events within the VRCs – including potentially further sub-domains may exist for other steps in the virus cycle such as transcription or virus assembly.
3D-SIM Microscopy Set-up
The 3D- SIM structured illumination images were acquired on a Nikon structured illumination microscope using a 1.49NA 100x oil SR Apo TIRF objective and illumination provided by 405/488/561/647 laser lines. An Andor iXon EMCCD camera was used as the imaging camera. The iXon EMCCD camera is an ideal solution for this technique as it is essential to have the most sensitive detector possible. EMCCD cameras are the most sensitive detector option due to the combination of high Quantum Efficiency and EM amplification that boosts even single photons detected to levels many time above the noise background. This provides better outright sensitivity than sCMOS cameras. The use of lower illumination intensities may also be possible, reducing phototoxicty and bleaching effects. The raw image data acquired was subsequently processed and reconstructed into super-resolved 3D-SIM images using the 3D-SIM module of NIS-Elements.
Research Paper: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008403
Featured Lab: Garcea Lab, University of Colorado
The Garcea laboratory studies the cell biology of small DNA viruses such as polyomavirus and papillomavirus. This includes characterization of viral assembly sites in the cell nucleus, and defining mitogenic signals resulting from the initial contact of the virus capsid with cell surface gangliosides. In addition virus infection studies , the laboratory is also investigating development potential vaccine candidates for human papillomavirus (HPV).
To find out more about the The Garcea Lab
Associated links
MCDB – Department of Molecular, Cellular, and Developmental Biology at the University of Colorado
BioFrontiers Institute – Colorado University’s interdisciplinary research facility
Light Microscopy Core Facility at University of Colorado Boulder
Selected Publications
- Justyna Broniarczyk, Paola Massimi, David Pim, Martina Bergant Marušič, Michael P. Myers, Robert L. Garcea, Lawrence Banks. Phosphorylation of Human Papillomavirus Type 16 L2 Contributes to Efficient Virus Infectious Entry. Journal of Virology Jun 2019, 93 (13) e00128-19; DOI:1128/JVI.00128-19
- Kimberly D. Erickson, Robert L. Garcea. Viral replication centers and the DNA damage response in JC virus-infected cells. Virology, Volume 528, 2019, Pages 198-206, ISSN 0042-6822. https://doi.org/10.1016/j.virol.2018.12.014
- Garcea, R.L., Meinerz, N.M., Dong, M. et al. Single-administration, thermostable human papillomavirus vaccines prepared with atomic layer deposition technology. npj Vaccines 5, 45 (2020). https://doi.org/10.1038/s41541-020-0195-4
- Katie Heiser, Catherine Nicholas, Robert L. Garcea. Activation of DNA damage repair pathways by murine polyomavirus. Virology, Volume 497, 2016, Pages 346-356, ISSN 0042-6822,. https://doi.org/10.1016/j.virol.2016.07.028.